Constructing Image DataSets: Difference between revisions
| Line 8: | Line 8: | ||
[[Image:unfold_image_graphic.png|Unfolding Images]] | [[Image:unfold_image_graphic.png|Unfolding Images]] | ||
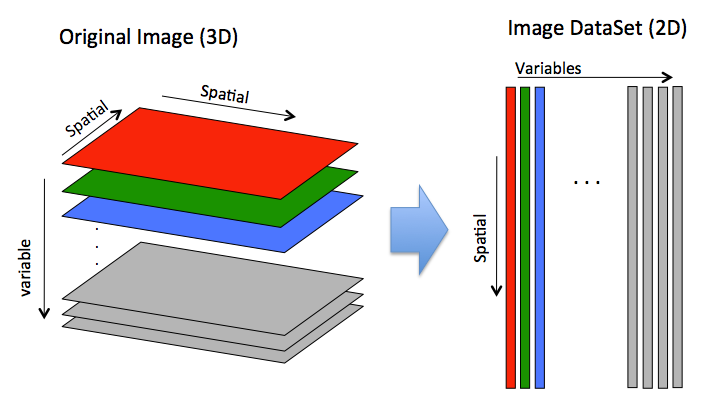
In the code below you can see a simple example image of random data (3px by 2px by 3 slabs/channels as if RBG) is created. The [[buildimage]] function is called to create an image DataSet. Each slab is "unfolded" (column wise) to become a column of raw data. Note the .imagesize | In the code below you can see a simple example image of random data (3px by 2px by 3 slabs/channels as if RBG) is created. The [[buildimage]] function is called to create an image DataSet. Each slab is "unfolded" (column wise) to become a column of raw data. Note the .imagesize field indicates the original size of the image and thus how to "fold" the data back into an image. | ||
<pre> | <pre> | ||
| Line 44: | Line 44: | ||
0.5076 0.9748 0.8915 | 0.5076 0.9748 0.8915 | ||
0.2380 0.5751 0.2140 | 0.2380 0.5751 0.2140 | ||
>> b.imagesize | |||
ans = | |||
3 2 | |||
</pre> | </pre> | ||
Revision as of 10:28, 22 May 2020
Building image DSOs can be accomplished via a GUI in Solo+MIA or PLS_Toolbox and MIA_Toolbox, or from the MATLAB command line when using PLS_Toolbox and MIA_Toolbox. A list of available methods can be found here.
Getting Started
In general, images are stored "unfolded" in an image DataSet where all spacial dimensions are unfolded into mode 1 and variables are unfolded into mode 2. You can retrieve a "folded" image from an image DataSet using the .imagedata field.
In the code below you can see a simple example image of random data (3px by 2px by 3 slabs/channels as if RBG) is created. The buildimage function is called to create an image DataSet. Each slab is "unfolded" (column wise) to become a column of raw data. Note the .imagesize field indicates the original size of the image and thus how to "fold" the data back into an image.
>> a = rand(3,2,3)
a(:,:,1) =
0.1605 0.2914
0.7544 0.5076
0.7935 0.2380
a(:,:,2) =
0.1927 0.1959
0.8087 0.9748
0.0667 0.5751
a(:,:,3) =
0.5832 0.8787
0.9500 0.8915
0.1000 0.2140
>> b = buildimage(a);
>> b.data
ans =
0.1605 0.1927 0.5832
0.7544 0.8087 0.9500
0.7935 0.0667 0.1000
0.2914 0.1959 0.8787
0.5076 0.9748 0.8915
0.2380 0.5751 0.2140
>> b.imagesize
ans =
3 2
From a GUI
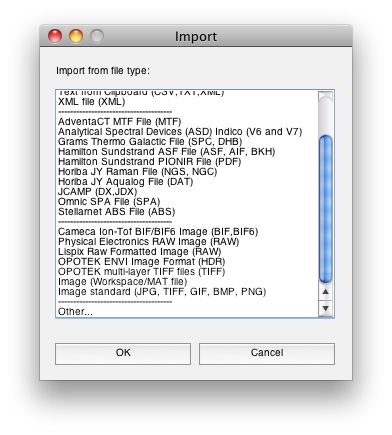
When using Solo+MIA or when you have MIA_Toolbox installed with PLS_Toolbox, image importing options automatically become available from the File/Import menus. You'll notice extra selections in the Import menu for images: File/Import Data/X Block. In this example we’re loading the JPEG from above directly into Analysis GUI.
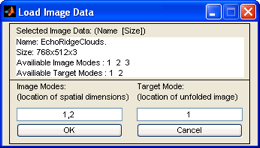
After you've selected the file you wish to import, you're prompted to specify what modes are spatial (m-by-n pixels) and which mode you'd like them unfolded to.
Since the spatial data resides in the first two modes (768x512 pixels) we enter "1,2" in
the Image Modes text box. Generally you put unfolded data into mode 1 of the new
image dataset, so we'll use the default value of "1" in the Target Mode text box. Clicking
OK loads the data into Analysis.
If you build a PCA model on this data you'll notice two things: an extra button on the toolbar, and different behavior when viewing data or scores.
The extra button opens the imagegui tool, see the MIA_Toolbox documentation for more information how imagegui works (or, if using PLS_Toolbox, type help imagegui at the command line). Also note that imagegui needs a model with at least 2 PCs calculated to be enabled.
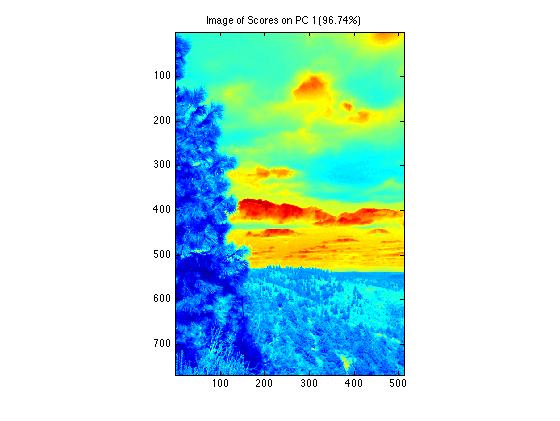
Viewing data and scores will display information in its "folded", image form. In this example the 2 pc model scores plot displays in the original image spatial dimensions.
Most methods in Analysis GUI are image “enabled” in that they will automatically work on image data (PARAFAC will still operate on images but won't display "folded" image data). Be aware of size involved in image data. Large volumes of data will require extended amounts of time and memory to analyze. When a dataset is too large it will generate out of memory errors.
Converting an Existing DataSet
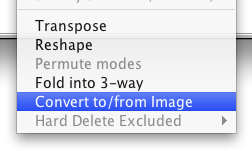
If you have an existing DataSet that is already unfolded, you can transform it into an image DataSet by defining the image size. This can be done from the DataSet Editor Transform menu (Transform>Convert to/from Image).
You will be prompted for information about the image. The first dialog box asks if the image is multivariate or univariate ( get more information about image types here). If you have a multivariate image you will then be prompted for the image size. This should be the size, in pixels of the original image. Once you've input the image size the function will then add this information to the DataSet and change it's type to "image".
From the MATLAB Command Line
With PLS_Toolbox and MIA_Toolbox, the easiest way to import an image via the command line is simply to use imageload. Calling imageload with no imputs will allow you to import and transform an image in the MATLAB workspace or from a .mat file.
imageload
Calling imageload with "other" switch will allow you to import from a number of standard image file types (.tiff, .png, .hdf, .bmp, .jpeg, .gif):
imageload('other')
The following example shows how to build up an image dataset "manually". It uses the EchoRidgeClouds.jpeg image from the MIA_Toolbox/dems folder. This JPEG image contains RGB (Truecolor) information of size N x M x 3 where N and M are the pixel (spatial) dimensions. The following commands will load the image using the MATLAB image read command imread then unfold the image into the first mode of an image dataset using the buildimage command.
>> myimage = imread('EchoRidgeClouds.jpeg');
>> myimage = buildimage(myimage ,[1 2], 1, 'myNewImage')
myimage =
name: myNewImage
type: image
author:
date: 03-Oct-2005 10:31:03
moddate: 03-Oct-2005 10:31:03
data: 393216x3 [double]
imagesize: 768x512
imagemode: 1
label: {2x1} [array (char)]
Mode 1 [: ]
Mode 2 [: ]
axisscale: {2x1} [vector (real)]
...
What if you have an existing DSO with image data in it? Use the exact same command:
>> myimage = buildimage(mydso ,[1 2], 1, 'myNewImage')
The last input to buildimage, 'myNewImage', renames the dataset (the default name is the variable name). The default setting for buildimage is to handle 3 dimensional data (three modes), see the documentation for specific settings.
Concatenating Images
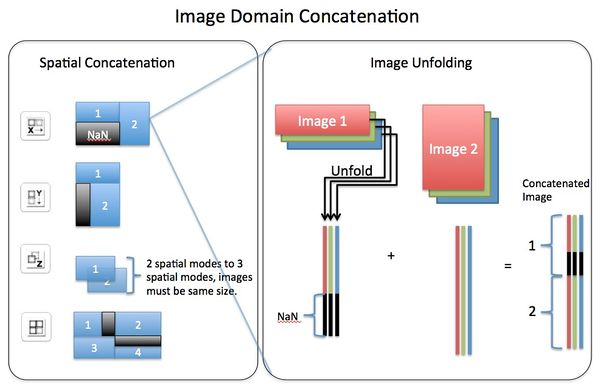
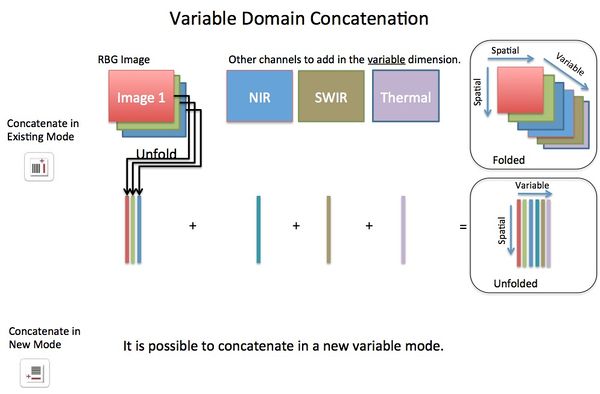
Concatenating images can be performed, via Image Manager, in either the image (X,Y pixel or spatial) domain or the variable (spectral) domain. In the spatial domain images can be concatenated in the X direction, Y direction, Z direction, or "tiled" in X/Y plane. Concatenation in the variable domain is almost always done in the "existing" variable mode (dimension). If concatenating in a new variable mode this will effectively create n-way data where data will be unfolded on the spatial modes but retain the variable modes.
NOTE: If images are concatenated in the Z direction this results in a 4 dimensional dataset that is pixels by pixels by pixels by variable. This is often confused with concatenating in the existing variable dimension where each image is "stacked" in the variable dimension.
NOTE: When concatenating in image (spatial) domain, a new class set will be created with each (sub) image assigned to a class. This will allow for class based preprocessing (e.g., class centering to remove offset).
Visual explanation of Image Domain Concatenation