Xgb: Difference between revisions
imported>Donal |
No edit summary |
||
| Line 8: | Line 8: | ||
:pred = xgb(x,model,options); %makes predictions with a new X-block | :pred = xgb(x,model,options); %makes predictions with a new X-block | ||
:valid = xgb(x,y,model,options); %performs a "test" call with a new X-block and known y-values | :valid = xgb(x,y,model,options); %performs a "test" call with a new X-block and known y-values | ||
Please note that the recommended way to build a Gradient Boosted Tree Ensemble for regression using XGBoost model from the command line is to use the Model Object. Please see [[EVRIModel_Objects | this wiki page on building models using the Model Object]]. | |||
===Description=== | ===Description=== | ||
| Line 70: | Line 72: | ||
===See Also=== | ===See Also=== | ||
[[analysis]], [[browse]], [[knn]], [[lwr]], [[pls]], [[plsda]], [[xgbda]], [[xgbengine]] | [[analysis]], [[browse]], [[knn]], [[lwr]], [[pls]], [[plsda]], [[xgbda]], [[xgbengine]], [[EVRIModel_Objects]] | ||
Revision as of 15:40, 3 January 2020
Purpose
Gradient Boosted Tree Ensemble for regression using XGBoost.
Synopsis
- model = xgb(x,y,options); %identifies model (calibration step)
- pred = xgb(x,model,options); %makes predictions with a new X-block
- valid = xgb(x,y,model,options); %performs a "test" call with a new X-block and known y-values
Please note that the recommended way to build a Gradient Boosted Tree Ensemble for regression using XGBoost model from the command line is to use the Model Object. Please see this wiki page on building models using the Model Object.
Description
To choose between regression and classification, use the xgbtype option:
- regression : xgbtype = 'xgbr'
- classification : xgbtype = 'xgbc'
It is recommended that classification be done through the xgbda function.
Inputs
- x = X-block (predictor block) class "double" or "dataset",
- y = Y-block (predicted block) class "double" or "dataset",
- model = previously generated model (when applying model to new data)
Outputs
- model = standard model structure containing the xgboost model (see Standard Model Structure). Feature scores are contained in model.detail.xgb.featurescores.
- pred = structure array with predictions
- valid = structure array with predictions
Options
options = a structure array with the following fields:
- display: [ 'off' | {'on'} ] governs level of display to command window.
- plots [ 'none' | {'final'} ] governs level of plotting.
- waitbar: [ off | {'on'} ] governs display of waitbar during optimization and predictions.
- preprocessing: {[] []}, two element cell array containing preprocessing structures (see PREPROCESS) defining preprocessing to use on the x- and y-blocks (first and second elements respectively)
- algorithm: [ 'xgboost' ] algorithm to use. xgboost is default and currently only option.
- classset : [ 1 ] indicates which class set in x to use when no y-block is provided.
- xgbtype : [ {'xgbr'} | 'xgbc' ] Type of XGB to apply. Default is 'xgbc' for classification, and 'xgbr' for regression.
- compression : [{'none'}| 'pca' | 'pls' ] type of data compression to perform on the x-block prior to calculaing or applying the XGB model. 'pca' uses a simple PCA model to compress the information. 'pls' uses either a pls or plsda model (depending on the xgbtype). Compression can make the XGB more stable and less prone to overfitting.
- compressncomp : [ 1 ] Number of latent variables (or principal components to include in the compression model.
- compressmd : [ 'no' |{'yes'}] Use Mahalnobis Distance correctedscores from compression model.
- cvi : { { 'rnd' 5 } } Standard cross-validation cell (see crossval)defining a split method, number of splits, and number of iterations. This cross-validation is use both for parameter optimization and for error estimate on the final selected parameter values.Alternatively, can be a vector with the same number of elements as x has rows with integer values indicating CV subsets (see crossval).
- eta : Value(s) to use for XGBoost 'eta' parameter. Eta controls the learning rate of the gradient boosting.Values in range (0,1]. Using a single value specifies the value to use. Using a range of values specifies the parameters to search over to find the optimal value. Default is 3 values [0.1, 0.3, 0.5].
- max_depth : Value(s) to use for XGBoost 'max_depth' parameter. Specifies the maximum depth allowed for the decision trees. Using a single value specifies the value to use. Using a range of values specifies the parameters to search over to find the optimal value. Default is 6 values [1 2 3 4 5 6].
- num_round : Value(s) to use for XGBoost 'num_round' parameter. Specifies how many rounds of tree creation to perform. Using a single value specifies the value to use. Using a range of values specifies the parameters to search over to find the optimal value. Default is 3 values [100 300 500].
Algorithm
Xgb is implemented using the XGBoost package. User-specified values are used for XGBoost parameters (see options above). See XGBoost Parameters for further details of these options.
The default XGB parameters eta, max_depth and num_round have value ranges rather than single values. This xgb function uses a search over the grid of appropriate parameters using cross-validation to select the optimal XGBoost parameter values and builds an XGB model using those values. This is the recommended usage. The user can avoid this grid-search by passing in single values for these parameters, however.
Choosing the best XGB parameters
The recommended technique is to repeatedly test XGB using different parameter values and select the parameter combination which gives the best results. XGB searches over ranges of parameters eta, max_depth, and num_round, by default. The actual values tested can be specified by the user by setting the associated parameter option value. Each test builds an XGB model on the calibration data using cross-validation to produce root mean square error (RMSECV) result for that test. These tests are compared over all tested parameter combinations to find which combination gives the best cross-validation prediction (smallest RMSECV). The XGB model is then built using the optimal parameter setting.
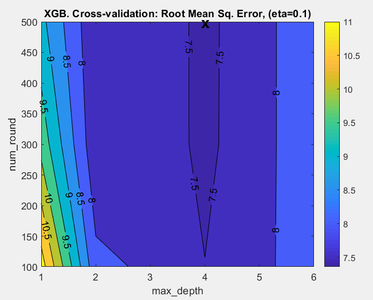
XGB parameter search summary plot
When XGB is run in the Analysis window it is possible to view the results of the XGB parameter search by clicking on the "Variance Captured" plot icon in the toolbar. If at least two XGB parameters were initialized with parameter ranges, for example eta and max_depth, then a figure appears showing the performance of the model plotted against eta and max_depth (Fig. 1). The measure of performance used is the root mean square error based on the cross-validation predictions predictions for the calibration data (RMSECV). The lowest value of RMSECV is marked on the plot by an "X" and this indicates the values of the XGB eta and max_depth parameters which yield the best performing model. The actual XGB model is built using these parameter values. If all three parameters, eta, max_depth, and num_round have ranges of values then you can view the prediction performance over the other variables' ranges by clicking on the blue horizontal arrow toolbar icon above the plot. In Analysis XGB the optimal parameters are also reported in the model summary window which is shown when you mouse-over the model icon, once the model is built. If you are using the command line XGB function to build a model then the optimal XGB parameters are shown in model.detail.xgb.cvscan.best.
- Fig. 1. Parameter search summary
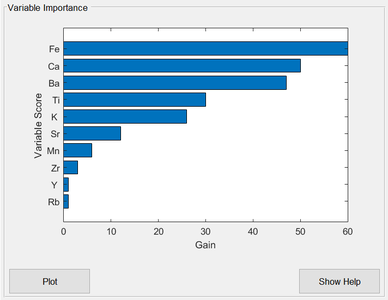
Variable Importance Plot
The ease of interpreting single decision trees is lost when a sequence of boosted trees is used, as in XGBoost. One commonly used diagnostic quantity for interpreting boosted trees is the "feature importance", or "variable importance" in PLS_Toolbox terminology. This is a measure of each variable's importance to the tree ensemble construction. It is calculated for each variable by summing up the “gain” on each node where that variable was used for splitting, over all trees in the sequence. "gain" refers to the reduction in the loss function being optimized. The important variables are shown in the XGB Analysis window when the model is built, ranked by their importance (Fig. 2).
- Fig. 2. Variable importance plot
See Also
analysis, browse, knn, lwr, pls, plsda, xgbda, xgbengine, EVRIModel_Objects