Gcluster: Difference between revisions
Jump to navigation
Jump to search
imported>Jeremy (Importing text file) |
imported>Benjamin No edit summary |
||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
===Purpose=== | ===Purpose=== | ||
Internal function used with cluster analysis in the [[Analysis_GUI|Analysis interface]]. | |||
===Synopsis=== | ===Synopsis=== | ||
: | :n/a | ||
===Description=== | ===Description=== | ||
This function is no longer an entry point for the clustering interface. To access a graphical user interface for cluster analysis, see the [[Analysis_GUI|Analysis interface]]. | |||
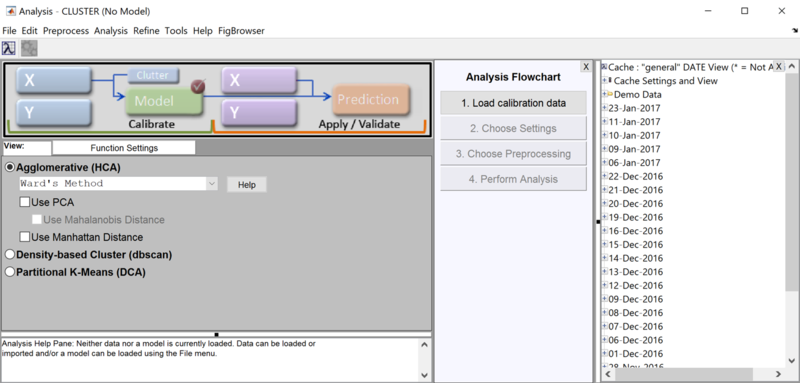
As of PLS_Toolbox/Solo 8.3, The gcluster GUI was updated to allow the use of Manhattan distance instead of Euclidean distance (default). Note the three checkboxes 'Use PCA', 'Use Mahalanobis Distance', and 'Use Manhattan Distance'. | |||
[[Image:gcluster_gui_manhattan.png|800px| Cluster in the Analysis GUI]] | |||
To keep the use of PCA scores (instead of the raw data) and the use of Manhattan Distance mutually exclusive, the following behaviors have been added to the GUI: | |||
:*If 'Use PCA' is checked: | |||
::* 'Use Mahalaobis Distance' will be enabled (to allow the user to check it). | |||
::* 'Use Manhattan Distance' will be unchecked. | |||
:*If 'Use Manhattan Distance' is checked: | |||
::* 'Use PCA will be unchecked'. | |||
::* 'Use Mahalanobis Distance' will be both unchecked and disabled. | |||
===See Also=== | ===See Also=== | ||
[[cluster]], [[simca]] | [[analysis]], [[analysis_GUI]], [[cluster]], [[pca]], [[simca]], [[manhattandist]] | ||
Latest revision as of 12:38, 16 August 2017
Purpose
Internal function used with cluster analysis in the Analysis interface.
Synopsis
- n/a
Description
This function is no longer an entry point for the clustering interface. To access a graphical user interface for cluster analysis, see the Analysis interface.
As of PLS_Toolbox/Solo 8.3, The gcluster GUI was updated to allow the use of Manhattan distance instead of Euclidean distance (default). Note the three checkboxes 'Use PCA', 'Use Mahalanobis Distance', and 'Use Manhattan Distance'.
To keep the use of PCA scores (instead of the raw data) and the use of Manhattan Distance mutually exclusive, the following behaviors have been added to the GUI:
- If 'Use PCA' is checked:
- 'Use Mahalaobis Distance' will be enabled (to allow the user to check it).
- 'Use Manhattan Distance' will be unchecked.
- If 'Use Manhattan Distance' is checked:
- 'Use PCA will be unchecked'.
- 'Use Mahalanobis Distance' will be both unchecked and disabled.