SpectraQuant Filtering
CPSA is a preprocessing method used to remove interferences from data prior to building a regression model. The CPSA filter includes one or more of the following corrections:
- multivariate filtering of components defined by a set of clutter or "correction" spectra.
- detrending (aka baselining) of the input data in windows of variables.
- an integrated thickness/pathlength correction.
CPSA Panel
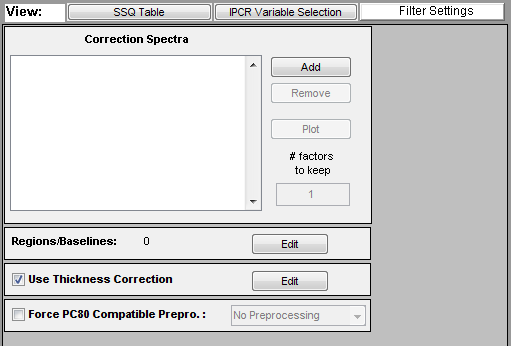
The CPSA tools are accessed through the CPSA panel of Analysis. This panel is only available when using the PCR or PLS Analysis methods and only enabled when calibration data has already been loaded. The default settings for the panel are shown below.
Four different options categories are available on the panel:
- Correction Spectra - Allows selecting correction spectra which describe background components which should be removed from the spectra prior to analysis.
- Regions/Baselines - Defines regions of the spectra and the baseline order to apply to each region.
- Thickness Correction - Determines whether or not to use thickness/pathlength correction (requires thickness/pathlength reference in calibration data)
- Force PC80 Compatible Preprocessing - Determines whether to enforce strict preprocessing rules on the created model.
Correction Spectra
The list on the left of this option shows the file(s) which have been selected by the user as background "clutter" spectra which should be removed from all calibration and test spectra (as well as future unknown spectra).
Adding Correction Spectra
To add spectra to the list, click on the Add button and select the file type to read (e.g. "Hamilton Sundstrand ASF files") and click OK.
Next, browse to the folder containing the file(s) to load, select one or more files (using shift-click or control-click to select multiple files), and click "Open".
The files will be loaded and added to the list. You will be advised if the spectral axis of the loaded files appears to be incompatible with the currently loaded x-block data.
Removing Correction Spectra
Force PC80 Compatible Preprocessing
If this option is unchecked, the user may use any preprocessing available in the custom preprocesing interface. However, the model created will be compatible with Eigenvector Research software such as Solo_Predictor, Solo, and PLS_Toolbox but will not be compatible with the Hamilton Sundstrand PC80 and CPSA32 applications.
When checked, the preprocessing selected in the pull-down menu to the right of the option (including: None, Mean Centering, Autoscaling) will be used and any other preprocessing will be removed. This mode assures that the model created will be as compatible as possible with the PC80 and CPSA32 applications.