Advanced Preprocessing: Sample Normalization
Introduction
In many analytical methods, the variables measured for a given sample are subject to overall scaling or gain effects. That is, all (or maybe just a portion of) the variables measured for a given sample are increased or decreased from their true value by a multiplicative factor. Each sample can, in these situations, experience a different level of multiplicative scaling.
In spectroscopic applications, scaling differences arise from pathlength effects, scattering effects, source or detector variations, or other general instrumental sensitivity effects (see, for example, Martens and Næs, 1989). Similar effects can be seen in other measurement systems due to physical or chemical effects (e.g., decreased activity of a contrast reagent or physical positioning of a sample relative to a sensor). In these cases, it is often the relative value of variables which should be used when doing multivariate modeling rather than the absolute measured value. Essentially, one attempts to use an internal standard or other pseudo-constant reference value to correct for the scaling effects.
The sample normalization preprocessing methods attempt to correct for these kinds of effects by identifying some aspect of each sample which should be essentially constant from one sample to the next, and correcting the scaling of all variables based on this characteristic. The ability of a normalization method to correct for multiplicative effects depends on how well one can separate the scaling effects which are due to properties of interest (e.g., concentration) from the interfering systematic effects.
Normalization also helps give all samples an equal impact on the model. Without normalization, some samples may have such severe multiplicative scaling effects that they will not be significant contributors to the variance and, as a result, will not be considered important by many multivariate techniques.
When creating discriminant analysis models such as PLS-DA or SIMCA models, normalization is done if the relationship between variables, and not the absolute magnitude of the response, is the most important aspect of the data for identifying a species (e.g., the concentration of a chemical isn't important, just the fact that it is there in a detectable quantity). Use of normalization in these conditions should be considered after evaluating how the variables' response changes for the different classes of interest. Models with and without normalization should be compared.
Typically, normalization should be performed before any centering or scaling or other column-wise preprocessing steps and after baseline or offset removal (see above regarding these preprocessing methods). The presence of baseline or offsets can impede correction of multiplicative effects. The effect of normalization prior to background removal will, in these cases, not improve model results and may deteriorate model performance.
One exception to this guideline of preprocessing order is when the baseline or background is very consistent from sample to sample and, therefore, provides a very useful reference for normalization. This can sometimes be seen in near-infrared spectroscopy in the cases where the overall background shape is due to general solvent or matrix vibrations. In these cases, normalization before background subtraction may provide improved models. In any case, cross-validation results can be compared for models with the normalization and background removal steps in either order and the best selected.
A second exception is when normalization is used after a scaling step (such as autoscaling). This should be used when autoscaling emphasizes features which may be useful in normalization. This reversal of normalization and scaling is sometimes done in discriminant analysis applications.
Normalize
Simple normalization of each sample is a common approach to the multiplicative scaling problem. The Normalize preprocessing method calculates one of several different metrics using all the variables of each sample. Possibilities include:
| Name | Description | Equation* |
|---|---|---|
| 1-Norm |
Normalize to (divide each variable by) the sum of the absolute value of all variables for the given sample. Returns a vector with unit area (area = 1) "under the curve." |
|
| 2-Norm |
Normalize to the sum of the squared value of all variables for the given sample. Returns a vector of unit length (length = 1). A form of weighted normalization where larger values are weighted more heavily in the scaling. |
|
| Inf-Norm |
Normalize to the maximum value observed for all variables for the given sample. Returns a vector with unit maximum value. Weighted normalization where only the largest value is considered in the scaling. |
- Where, in each case, wi is the normalization weight for sample i, xi is the vector of observed values for the given sample, j is the variable number, and n is the total number of variables (columns of x).
The weight calculated for a given sample is then used to calculate the normalized sample, , using:
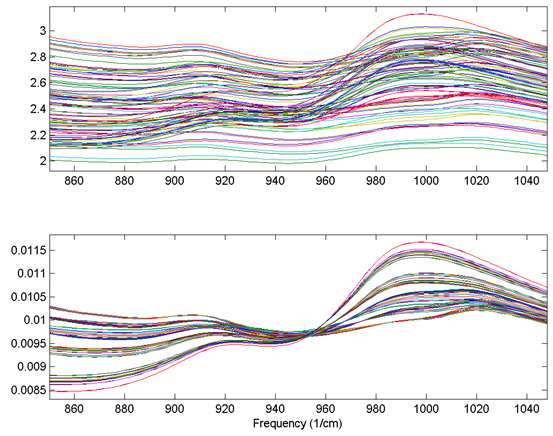
An example using the 1-norm on near infrared spectra is shown in the figure below. These spectra were measured as 20 replicates of 5 synthetic mixtures of gluten and starch (Martens, Nielsen, and Engelsen, 2003) In the original data (top plot), the five concentrations of gluten and starch are not discernable because of multiplicative and baseline effects among the 20 replicate measurements of each mixture. After normalization using a 1-norm (bottom plot), the five mixtures are clearly observed in groups of 20 replicate measurements each.
 Normal 0 false false false EN-US X-NONE X-NONE MicrosoftInternetExplorer4
Normal 0 false false false EN-US X-NONE X-NONE MicrosoftInternetExplorer4
Figure: Effect of normalization on near-IR spectra of five synthetic gluten and starch mixtures. Original spectra (top plot) and spectra after 1-norm normalization (bottom plot) are shown.
From the Preprocessing GUI, the only setting associated with this method is the type of normalization (1-norm, 2-norm or inf-norm). There is currently no option to perform this normalization based on anything other than all selected variables.
From the command line, this method is performed using the normaliz function (note the unusual spelling of the function).
SNV (Standard Normal Variate)
Unlike the simple 1-Norm Normalize described above, the Standard Normal Variate (SNV) normalization method is a weighted normalization (i.e., not all points contribute to the normalization equally). SNV calculates the standard deviation of all the pooled variables for the given sample (see for example Barnes et al., 1989). The entire sample is then normalized by this value, thus giving the sample a unit standard deviation (s = 1). Note that this procedure also includes a zero-order detrend (subtraction of the individual mean value from each spectrum - see discussion of detrending, above), and also that this is different from mean centering (described later). The equations used by the algorithm are the mean and standard deviation equations:





