Applying a Model Quick Start: Difference between revisions
imported>Jeremy No edit summary |
imported>Scott No edit summary |
||
| (14 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[Review_Results_Quick_Start | Previous Topic: Review Results]] | |||
[[ | |||
* '' | __TOC__ | ||
==Load the Model== | |||
Congratulations! You have collected calibration data and gone through the exercise of building a model that meets your objectives. Now, you want to exert one of the most stringent tests - applying your model to new data. If you have just completed the model building process, all that needs to be done is to load some new data as validation data. Another scenario is that you have a model that has been built awhile ago, and you wish to apply it to some new data. | |||
In this example (using the included demonstration dataset "'''''nir_data'''''") , there are three variables in the workspace | |||
* ''pls_model...'' - a PLS model that has been built on spectral data (''spec1'') to predict a concentration | |||
* ''spec2'' - a new set of spectral data to be used to validate the model | * ''spec2'' - a new set of spectral data to be used to validate the model | ||
* ''conc'' - concentration data for the validation spectra | * ''conc'' - concentration data for the validation spectra | ||
The concentration data contains values for five separate components. The model '' | The concentration data contains values for five separate components. The model ''pls_model...'' predicts only one of these concentration values. | ||
To load the model and data into the workspace: | |||
* In the Model Cache panel (of the Workspace Browser), double-click the model. | |||
* Double-click the demo data set NIR (under the Demo Data section). | |||
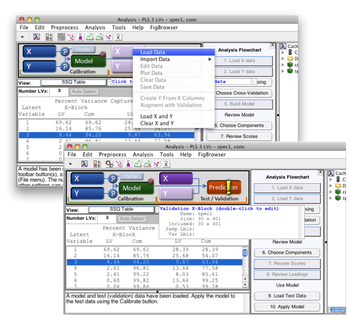
* In the Workspace panel, double-click the model, this will open a new Analysis window and load the model. You will see the SSQ table populated with values, indicating that the model has been loaded. If the model cache was activated during the course of building the model and remains so, the calibration data will also be loaded. You can see this by noting that the '''X''' and '''Y''' buttons appear depressed, and when you pass the mouse cursor over either information on the respective data blocks is revealed. | |||
[[Image:apply_model.013.png| |500px|(Click to Enlarge)]] | |||
==Load Test Data and Apply Model== | |||
Now that the model has been loaded | Now that the model has been loaded | ||
* click | * Right click on the Validation X icon and select Load Data. | ||
* Click on the '''"Apply Model"''' button under the Analysis Flowchart | |||
* | |||
[[Image:LoadValX.png| |350px|(Click to Enlarge)]] | |||
==Review Results== | |||
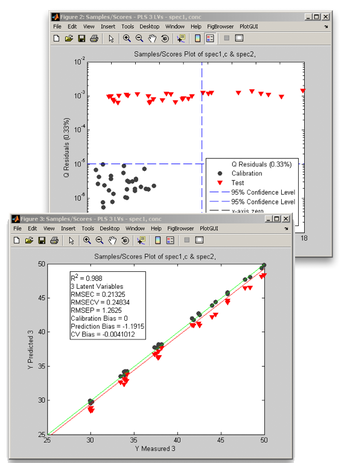
When you click on the "Review Scores" button, a multiplot figure will open. Try double-clicking on each subplot to create separate figures. One useful plot is Q residuals versus Hotelling's T<sup>2</sup>, with both the validation and calibration data visible. In the Plot Controls window, select "Hotelling T^2" for the x-axis, and "Q Residuals" for the y-axis. Make sure that the "Show Cal Data with Test" box is checked toward the bottom of the Plot Controls window. Finally, select "View" under the Plot Controls menu, then "Classes", followed by "Cal/Test Samples"; this will apply color/symbol coding for the two classes of samples. It is sometimes useful to use log scales for Q residuals and/or T<sup>2</sup>; in this example, a log scale is used for the Q residuals. For the second plot, "Y Predicted" is selected for the y-axis along with "Y Measured" for the x-axis. As in the Q residuals vs. T<sup>2</sup> plot, the black circles represent the calibration samples and the red triangles denote the validation samples. | When you click on the "Review Scores" button, a multiplot figure will open. Try double-clicking on each subplot to create separate figures. One useful plot is Q residuals versus Hotelling's T<sup>2</sup>, with both the validation and calibration data visible. In the Plot Controls window, select "Hotelling T^2" for the x-axis, and "Q Residuals" for the y-axis. Make sure that the "Show Cal Data with Test" box is checked toward the bottom of the Plot Controls window. Finally, select "View" under the Plot Controls menu, then "Classes", followed by "Cal/Test Samples"; this will apply color/symbol coding for the two classes of samples. It is sometimes useful to use log scales for Q residuals and/or T<sup>2</sup>; in this example, a log scale is used for the Q residuals. For the second plot, "Y Predicted" is selected for the y-axis along with "Y Measured" for the x-axis. As in the Q residuals vs. T<sup>2</sup> plot, the black circles represent the calibration samples and the red triangles denote the validation samples. | ||
| Line 33: | Line 42: | ||
* a suggested step would be to determine what are the factors that contribute to the high values of Q residuals and T<sup>2</sup>; these are readily obtained by using the '''Q con''' and '''T con''' buttons on the Plot Controls window | * a suggested step would be to determine what are the factors that contribute to the high values of Q residuals and T<sup>2</sup>; these are readily obtained by using the '''Q con''' and '''T con''' buttons on the Plot Controls window | ||
[[Image:apply_model.015.png| |350px|(Click to Enlarge)]] | |||
Latest revision as of 10:04, 22 September 2011
Previous Topic: Review Results
Load the Model
Congratulations! You have collected calibration data and gone through the exercise of building a model that meets your objectives. Now, you want to exert one of the most stringent tests - applying your model to new data. If you have just completed the model building process, all that needs to be done is to load some new data as validation data. Another scenario is that you have a model that has been built awhile ago, and you wish to apply it to some new data.
In this example (using the included demonstration dataset "nir_data") , there are three variables in the workspace
- pls_model... - a PLS model that has been built on spectral data (spec1) to predict a concentration
- spec2 - a new set of spectral data to be used to validate the model
- conc - concentration data for the validation spectra
The concentration data contains values for five separate components. The model pls_model... predicts only one of these concentration values.
To load the model and data into the workspace:
- In the Model Cache panel (of the Workspace Browser), double-click the model.
- Double-click the demo data set NIR (under the Demo Data section).
- In the Workspace panel, double-click the model, this will open a new Analysis window and load the model. You will see the SSQ table populated with values, indicating that the model has been loaded. If the model cache was activated during the course of building the model and remains so, the calibration data will also be loaded. You can see this by noting that the X and Y buttons appear depressed, and when you pass the mouse cursor over either information on the respective data blocks is revealed.
Load Test Data and Apply Model
Now that the model has been loaded
- Right click on the Validation X icon and select Load Data.
- Click on the "Apply Model" button under the Analysis Flowchart
Review Results
When you click on the "Review Scores" button, a multiplot figure will open. Try double-clicking on each subplot to create separate figures. One useful plot is Q residuals versus Hotelling's T2, with both the validation and calibration data visible. In the Plot Controls window, select "Hotelling T^2" for the x-axis, and "Q Residuals" for the y-axis. Make sure that the "Show Cal Data with Test" box is checked toward the bottom of the Plot Controls window. Finally, select "View" under the Plot Controls menu, then "Classes", followed by "Cal/Test Samples"; this will apply color/symbol coding for the two classes of samples. It is sometimes useful to use log scales for Q residuals and/or T2; in this example, a log scale is used for the Q residuals. For the second plot, "Y Predicted" is selected for the y-axis along with "Y Measured" for the x-axis. As in the Q residuals vs. T2 plot, the black circles represent the calibration samples and the red triangles denote the validation samples.
We note from these two plots that:
- the validation samples are markedly different from the calibration samples - there is at least a two order of magnitude difference in Q residuals between the validation and calibration sets, and several of the validation samples have values of T2 that are higher than the 95% confidence limit
- the predictions for the validation samples are biased to lower values, although the correlation as measured by R2 is still high
- a suggested step would be to determine what are the factors that contribute to the high values of Q residuals and T2; these are readily obtained by using the Q con and T con buttons on the Plot Controls window