Demonstration Datasets: Difference between revisions
Jump to navigation
Jump to search
imported>Jeremy No edit summary |
imported>Lyle |
||
| (16 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
__TOC__ | |||
{| class="wikitable" | ==List of DataSets== | ||
The following is a list of the data supplied with PLS_Toolbox. It consists of the name of the file (all end in ".mat" unless otherwise specified) along with a brief description of the contents of the given file. Each file may contain one or more variables to be used. | |||
{| class="wikitable sortable" | |||
|- | |||
!File Name !! Description !! Variable Name(s) !! Variable Size !! Task !! Notes | |||
|- | |||
|alcohol || Biological fluid analysis of alcoholics || alcohol || 65x52 || Classification || | |||
|- | |||
|aminoacids || Fluorescence EEM of 5 aminoacid samples || X || 5x20x61 || Decomposition || Try with PARAFAC | |||
|- | |||
|arch || Archeological artifact data set || arch || 75x10 || Decomposition, Classification || | |||
|- | |||
|beer || VIS-NIR transmission recorded directly on undiluted degassed beer ||beer <br />extract <br />beertest <br />extracttest || 40x926 <br />40x1 <br />20x926 <br /> 20x1 ||Regression || Good for testing Variable Selection Interface | |||
|- | |||
|biscuit || NIR reflectance spectra of 40 samples of biscuit dough from 1200-2398nm. ||spec <br />recipe || 40x600 <br />40x4 || Regression || | |||
|- | |||
|brain_weight || Body mass (kg) and brain mass (g) for 28 animals. || brains || 28x2 || Decomposition || | |||
|- | |||
|bread || Sensory evaluation of breads. || bread || 10x11x8 || Decomposisiton || Try with PARAFAC, also good for testing EEM Filtering preprocessing | |||
|- | |||
|cancer || Fluorescence EEM spectra extracted from cervical cancer images. || cancer || 563x22 || Classification, Decomposition || Unfolded EEM data | |||
|- | |||
|corn_dso || 80 samples of corn measured on 3 different NIR spectrometers with moisture, oil, protein and starch values for each of the samples is also included. ||conc <br />m5nbs <br />m5spec <br />mp5nbs <br />mp5spec <br />mp6nbs <br />mp6spec || 80x4 <br />3x700 <br />80x700<br />4x700<br />80x700<br />4x700<br />80x700 || Regression || | |||
|- | |- | ||
| | |data_mid_IR || Data sets for correlation spectroscopy. || data_mid_IR || 21x130 || Correlation Spectroscopy || Use with data_near_IR dataset | ||
|- | |- | ||
| | |data_near_IR || Data sets for correlation spectroscopy. || data_near_IR || 21x149 || Correlation Spectroscopy || Use with data_mid_IR dataset | ||
|- | |- | ||
| | |dorrit || DORRIT EEM of 27 samples with 4 flourophores for PARAFAC. ||EEM <br />yblock || 27x116x18 <br /> 27x4 || Decomposition, Regression || Try PARAFAC and Multi-Way PLS, NPLS | ||
|- | |- | ||
| | |Dupont_BSPC || 10 process variables (pressure, flow, temperature) for 55 batches with 7 steps each. ||dupont_cal <br /> dupont_test || 3600x10 <br /> 1900x10 || Batch Processor || | ||
|- | |- | ||
| | |etchdata || Engineering process data from semiconductor metal etch ||Etchcal <br />EtchTest || 20x12x107 <br />20x12x20 || Decomposition || Use Multi-Way PCA (MPCA). See Chemometrics Tutorial chapter 5. | ||
|- | |- | ||
| | |fia || UV detection of Flow Injection Analysis of hydroxy-benzaldehyde (n-way data: sample, wavelength, time) || FIA || 12x50x45 || Decomposition || | ||
|- | |- | ||
| | |flucuttest || Fluorescence EEM data || z || 2x15x23 || Decomposition || Try PARAFAC | ||
|- | |- | ||
|FTIR_microscopy || FTIR microscopy transect spectra of a three-layer polymer laminate. | |FTIR_microscopy || FTIR microscopy transect spectra of a three-layer polymer laminate. || FTIR_microscopy || 17x81 || Decomposition || Use PURITY program | ||
|- | |- | ||
| | |gcwine || Dynamic headspace GCMS data of red wines from different regions. || gcwine || 71x150x24 || Decomposition || Try PARAFAC2 | ||
|- | |- | ||
| | |halddata || HALDDATA Hald cement curing data. ||xblock <br /> yblock || 13x4<br />13x1 || Regression || | ||
|- | |- | ||
| | |lcms || LC/MS electrospray of 15 surfactant solution. || lcms || 345x1451 || Decomposition || Use CODA-DW Tool. See Chemometrics Tutorial, chapter 9. | ||
|- | |- | ||
| | |lcms_compare1 || Select data from LC/MS electrospray data set. || lcms_compare1 || 675x1940 || Decomposition || Try LCMS Compare Tool. Use with lcms_compare2 and lcms_compare3. See Chemometris Tutorial Chapter, chapter 9. | ||
|- | |- | ||
| | |lcms_compare2 || Select data from LC/MS electrospray data set. || lcms_compare2 || 675x1940 || Decomposition || Try LCMS Compare Tool. Use with lcms_compare1 and lcms_compare3. See Chemometris Tutorial Chapter, chapter 9. | ||
|- | |- | ||
| | |lcms_compare3 || Select data from LC/MS electrospray data set. || lcms_compare3 || 675x1940 || Decomposition || Try LCMS Compare Tool. Use with lcms_compare1 and lcms_compare2. See Chemometris Tutorial Chapter, chapter 9. | ||
|- | |- | ||
| | |MS_time_resolved || Direct probe time profile MS of three color-coupling compounds. ||MS_time_resolved || 20x757 || Decomposition || Use PURITY program (try 3 and 4 components) | ||
|- | |- | ||
| | |MS_time_resolved_references || Reference spectra of pure compounds from MS_time_resolved. || MS_time_resolved_references || 3x757 || || Use along with MS_time_resolved data for comparing results from PURITY | ||
|- | |- | ||
| | |nir_data || NIR spectra of pseudo gasoline samples ||conc <br />spec1 <br />spec2 || 30x5 <br />30x401 <br />30x401 || Regression || Try Savitsky-Golay preprocessing | ||
|- | |- | ||
| | |nmr_data || NMR data for GRAM demo. || nmrdata || 20x1176 || Decomposition || | ||
|- | |- | ||
| | |octane || NIR spectra and octane number values of 39 gasoline samples. ||spec <br />octane || 39x226 <br />39x1 || Regression, Decomposition || Try some Robust methods. See Chemometrics Tutorial, chapter 13. | ||
|- | |- | ||
| | |oesdata || Optical emission spectra from metal etch. || oes1 || 46x770 || Decomposition || Try Multivariate Curve Resolution (MCR). See Chemometrics Tutorial, chapter 9. | ||
|- | |- | ||
| | |OliveOilData || 36 FT-IR spectra (3600 - 600 cm-1) of olive oils. ||xcal <br /> xtest || 36x518 <br /> 36x518 || Decomposition, Classification || Try Multiplicative Scatter Correction (MSC) preprocessing. | ||
|- | |- | ||
| | |paint || Non-linear paint formulation data.||paint_cal_X <br />paint_cal_Y <br />paint_test_X <br />paint_test_Y || 49x4<br />49x3 <br />8x4 <br />8x3 || Regression || Try using with non-linear regression methods, like SVM-R | ||
|- | |- | ||
| | |pcadata || Slurry Fed Ceramic Melter data. ||part1 <br />part2 || 300x10 <br />200x10 || Decomposition || | ||
|- | |- | ||
| | |plsdata || SFCM data for PCR and PLS demos. ||xblock1 <br />yblock1 <br />xblock2<br />yblock2 ||300x20 <br />300x1 <br />200x20 <br />200x1 || Regression || Try Multiple Linear Regression (MLR). See Chemometrics Tutorial chapter 6. | ||
|- | |- | ||
| | |pulsdata || Time series data from a Slurry Fed Ceramic Melter for PLSPULSM demo. || melter_data || 325x3 || Decomposition || | ||
|- | |- | ||
| | |purvardata || Raman spectra used in "Chemometrics Tutorial" section "MATLAB Code for Pure Variable Method, (chapter 9)". || data || 6x99 || Decomposition || Use PURITY | ||
|- | |- | ||
| | |purvardata_noise || Raman spectra used in "Chemometrics Tutorial" section "MATLAB Code for Pure Variable Method, (chapter 9)". || data || 6x99 || Decomposition || Use PURITY | ||
|- | |- | ||
| | |raman_dust_particles || Raman spectra || raman_dust_particles || 120x1025 || Decomposition || Use PURITY. Use with raman_dust_particles_references. See Chemometrics Tutorial section "MATLAB Code for Pure Variable Method, chapter 9. <br />Also try different Baseline preprocessing techniques with this data (Hint: order = 3). | ||
|- | |- | ||
| | |raman_dust_particles_references || Raman spectra || raman_dust_particles_references || 3x1025 || Decomposition || Use PURITY. Use with raman_dust_particles. See Chemometrics Tutorial section "MATLAB Code for Pure Variable Method, chapter 9 | ||
|- | |- | ||
| | |raman_time_resolved || Raman spectra of a time resolved reaction, used in "Chemometrics Tutorial". || raman_time_resolved || 16x151 || Decomposition || Try the PURITY program | ||
|- | |- | ||
| | |sawdata || Surface acoustic wave sensor data. || SAWdata || 72x13 || Decomposition, Classification || | ||
|- | |- | ||
| | |SBRdata_EU || NIR transmission spectra of styrene-butadiene copolymers. ||Xcal <br />Ycal <br />Xtest <br />Ytest || 60x141 <br />60x4 <br />10x141 <br />10x4 || Regression || Try different regression methods (CLS, MLR, PLS) | ||
|- | |- | ||
| | |stars || Surface temperature and light intensity values for 47 stars. || stars || 47x2 || Decomposition || | ||
|- | |||
|sugar || Sugar Fluorescence EEM N-way data set|| sugar || 268x44x7 || Decomposition || Try Mulit-Way PCA | |||
|- | |||
|wine || Wine demographic data set for PCA example. || wine || 10x5 || Decomposition || | |||
|- | |||
|wineregion || Metal Composition of Wines for classification by region. || wineregion || 38x17 || Classification || | |||
|} | |} | ||
==How to Load Demo Data== | |||
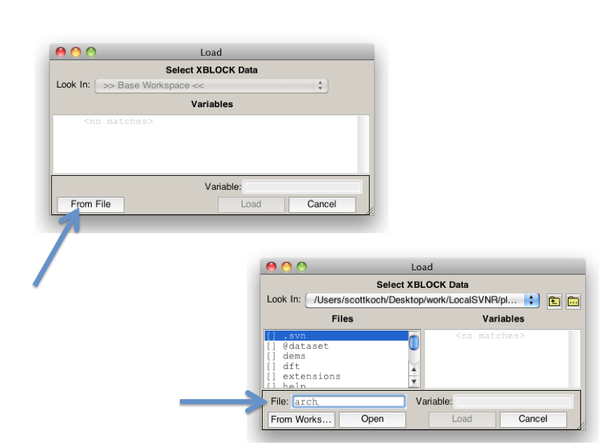
For MAT files, the easiest way to load demo data is using the Load Dialog Box: | |||
# From the file menu Load Data (or Import Workspace/MAT File). | |||
# When the dialog box appears click the From File button. | |||
# Type the name of the demo MAT file to loaded and hit the return key. | |||
# Select the variables to be loaded and click the Load button. | |||
[[Image:LoadDemoData.png| |600px|Load Demo Data]] | |||
(Sub topic of [[PLS_Toolbox_Topics|PLS_Toolbox_Topics]]) | (Sub topic of [[PLS_Toolbox_Topics|PLS_Toolbox_Topics]]) | ||
Latest revision as of 16:07, 8 May 2019
List of DataSets
The following is a list of the data supplied with PLS_Toolbox. It consists of the name of the file (all end in ".mat" unless otherwise specified) along with a brief description of the contents of the given file. Each file may contain one or more variables to be used.
| File Name | Description | Variable Name(s) | Variable Size | Task | Notes |
|---|---|---|---|---|---|
| alcohol | Biological fluid analysis of alcoholics | alcohol | 65x52 | Classification | |
| aminoacids | Fluorescence EEM of 5 aminoacid samples | X | 5x20x61 | Decomposition | Try with PARAFAC |
| arch | Archeological artifact data set | arch | 75x10 | Decomposition, Classification | |
| beer | VIS-NIR transmission recorded directly on undiluted degassed beer | beer extract beertest extracttest |
40x926 40x1 20x926 20x1 |
Regression | Good for testing Variable Selection Interface |
| biscuit | NIR reflectance spectra of 40 samples of biscuit dough from 1200-2398nm. | spec recipe |
40x600 40x4 |
Regression | |
| brain_weight | Body mass (kg) and brain mass (g) for 28 animals. | brains | 28x2 | Decomposition | |
| bread | Sensory evaluation of breads. | bread | 10x11x8 | Decomposisiton | Try with PARAFAC, also good for testing EEM Filtering preprocessing |
| cancer | Fluorescence EEM spectra extracted from cervical cancer images. | cancer | 563x22 | Classification, Decomposition | Unfolded EEM data |
| corn_dso | 80 samples of corn measured on 3 different NIR spectrometers with moisture, oil, protein and starch values for each of the samples is also included. | conc m5nbs m5spec mp5nbs mp5spec mp6nbs mp6spec |
80x4 3x700 80x700 4x700 80x700 4x700 80x700 |
Regression | |
| data_mid_IR | Data sets for correlation spectroscopy. | data_mid_IR | 21x130 | Correlation Spectroscopy | Use with data_near_IR dataset |
| data_near_IR | Data sets for correlation spectroscopy. | data_near_IR | 21x149 | Correlation Spectroscopy | Use with data_mid_IR dataset |
| dorrit | DORRIT EEM of 27 samples with 4 flourophores for PARAFAC. | EEM yblock |
27x116x18 27x4 |
Decomposition, Regression | Try PARAFAC and Multi-Way PLS, NPLS |
| Dupont_BSPC | 10 process variables (pressure, flow, temperature) for 55 batches with 7 steps each. | dupont_cal dupont_test |
3600x10 1900x10 |
Batch Processor | |
| etchdata | Engineering process data from semiconductor metal etch | Etchcal EtchTest |
20x12x107 20x12x20 |
Decomposition | Use Multi-Way PCA (MPCA). See Chemometrics Tutorial chapter 5. |
| fia | UV detection of Flow Injection Analysis of hydroxy-benzaldehyde (n-way data: sample, wavelength, time) | FIA | 12x50x45 | Decomposition | |
| flucuttest | Fluorescence EEM data | z | 2x15x23 | Decomposition | Try PARAFAC |
| FTIR_microscopy | FTIR microscopy transect spectra of a three-layer polymer laminate. | FTIR_microscopy | 17x81 | Decomposition | Use PURITY program |
| gcwine | Dynamic headspace GCMS data of red wines from different regions. | gcwine | 71x150x24 | Decomposition | Try PARAFAC2 |
| halddata | HALDDATA Hald cement curing data. | xblock yblock |
13x4 13x1 |
Regression | |
| lcms | LC/MS electrospray of 15 surfactant solution. | lcms | 345x1451 | Decomposition | Use CODA-DW Tool. See Chemometrics Tutorial, chapter 9. |
| lcms_compare1 | Select data from LC/MS electrospray data set. | lcms_compare1 | 675x1940 | Decomposition | Try LCMS Compare Tool. Use with lcms_compare2 and lcms_compare3. See Chemometris Tutorial Chapter, chapter 9. |
| lcms_compare2 | Select data from LC/MS electrospray data set. | lcms_compare2 | 675x1940 | Decomposition | Try LCMS Compare Tool. Use with lcms_compare1 and lcms_compare3. See Chemometris Tutorial Chapter, chapter 9. |
| lcms_compare3 | Select data from LC/MS electrospray data set. | lcms_compare3 | 675x1940 | Decomposition | Try LCMS Compare Tool. Use with lcms_compare1 and lcms_compare2. See Chemometris Tutorial Chapter, chapter 9. |
| MS_time_resolved | Direct probe time profile MS of three color-coupling compounds. | MS_time_resolved | 20x757 | Decomposition | Use PURITY program (try 3 and 4 components) |
| MS_time_resolved_references | Reference spectra of pure compounds from MS_time_resolved. | MS_time_resolved_references | 3x757 | Use along with MS_time_resolved data for comparing results from PURITY | |
| nir_data | NIR spectra of pseudo gasoline samples | conc spec1 spec2 |
30x5 30x401 30x401 |
Regression | Try Savitsky-Golay preprocessing |
| nmr_data | NMR data for GRAM demo. | nmrdata | 20x1176 | Decomposition | |
| octane | NIR spectra and octane number values of 39 gasoline samples. | spec octane |
39x226 39x1 |
Regression, Decomposition | Try some Robust methods. See Chemometrics Tutorial, chapter 13. |
| oesdata | Optical emission spectra from metal etch. | oes1 | 46x770 | Decomposition | Try Multivariate Curve Resolution (MCR). See Chemometrics Tutorial, chapter 9. |
| OliveOilData | 36 FT-IR spectra (3600 - 600 cm-1) of olive oils. | xcal xtest |
36x518 36x518 |
Decomposition, Classification | Try Multiplicative Scatter Correction (MSC) preprocessing. |
| paint | Non-linear paint formulation data. | paint_cal_X paint_cal_Y paint_test_X paint_test_Y |
49x4 49x3 8x4 8x3 |
Regression | Try using with non-linear regression methods, like SVM-R |
| pcadata | Slurry Fed Ceramic Melter data. | part1 part2 |
300x10 200x10 |
Decomposition | |
| plsdata | SFCM data for PCR and PLS demos. | xblock1 yblock1 xblock2 yblock2 |
300x20 300x1 200x20 200x1 |
Regression | Try Multiple Linear Regression (MLR). See Chemometrics Tutorial chapter 6. |
| pulsdata | Time series data from a Slurry Fed Ceramic Melter for PLSPULSM demo. | melter_data | 325x3 | Decomposition | |
| purvardata | Raman spectra used in "Chemometrics Tutorial" section "MATLAB Code for Pure Variable Method, (chapter 9)". | data | 6x99 | Decomposition | Use PURITY |
| purvardata_noise | Raman spectra used in "Chemometrics Tutorial" section "MATLAB Code for Pure Variable Method, (chapter 9)". | data | 6x99 | Decomposition | Use PURITY |
| raman_dust_particles | Raman spectra | raman_dust_particles | 120x1025 | Decomposition | Use PURITY. Use with raman_dust_particles_references. See Chemometrics Tutorial section "MATLAB Code for Pure Variable Method, chapter 9. Also try different Baseline preprocessing techniques with this data (Hint: order = 3). |
| raman_dust_particles_references | Raman spectra | raman_dust_particles_references | 3x1025 | Decomposition | Use PURITY. Use with raman_dust_particles. See Chemometrics Tutorial section "MATLAB Code for Pure Variable Method, chapter 9 |
| raman_time_resolved | Raman spectra of a time resolved reaction, used in "Chemometrics Tutorial". | raman_time_resolved | 16x151 | Decomposition | Try the PURITY program |
| sawdata | Surface acoustic wave sensor data. | SAWdata | 72x13 | Decomposition, Classification | |
| SBRdata_EU | NIR transmission spectra of styrene-butadiene copolymers. | Xcal Ycal Xtest Ytest |
60x141 60x4 10x141 10x4 |
Regression | Try different regression methods (CLS, MLR, PLS) |
| stars | Surface temperature and light intensity values for 47 stars. | stars | 47x2 | Decomposition | |
| sugar | Sugar Fluorescence EEM N-way data set | sugar | 268x44x7 | Decomposition | Try Mulit-Way PCA |
| wine | Wine demographic data set for PCA example. | wine | 10x5 | Decomposition | |
| wineregion | Metal Composition of Wines for classification by region. | wineregion | 38x17 | Classification |
How to Load Demo Data
For MAT files, the easiest way to load demo data is using the Load Dialog Box:
- From the file menu Load Data (or Import Workspace/MAT File).
- When the dialog box appears click the From File button.
- Type the name of the demo MAT file to loaded and hit the return key.
- Select the variables to be loaded and click the Load button.
(Sub topic of PLS_Toolbox_Topics)