Horibamultimodelpredictor: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| Line 27: | Line 27: | ||
* When a method has been assembled, use the right-click '''Validate Mehtod''' menu item to check that the sizes expected by the multiblock model are the same as those expected by the regression/classification models. | * When a method has been assembled, use the right-click '''Validate Mehtod''' menu item to check that the sizes expected by the multiblock model are the same as those expected by the regression/classification models. | ||
* To remove a method, mover or delete the .met file from the parent folder. | * To remove a method, mover or delete the .met file from the parent folder. | ||
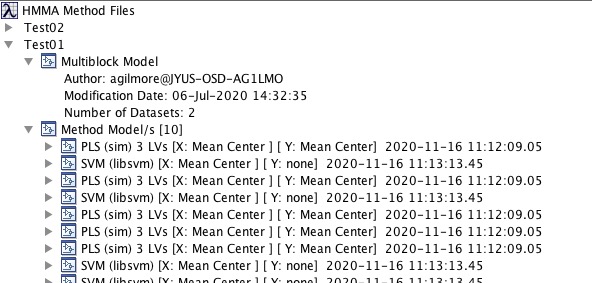
[[File:HMMAMethodEditTree.jpg| |HMMA Method Tree]] | |||
Revision as of 17:12, 2 February 2021
Multi-Model Predictor
Introduction
The Multi-Model Predictor is an Add-In component for Solo developed by HORIBA Scientific for use with HORIBA Fluorescence systems. To enable the Add-In contact visit www.horiba.com/fluorescence.
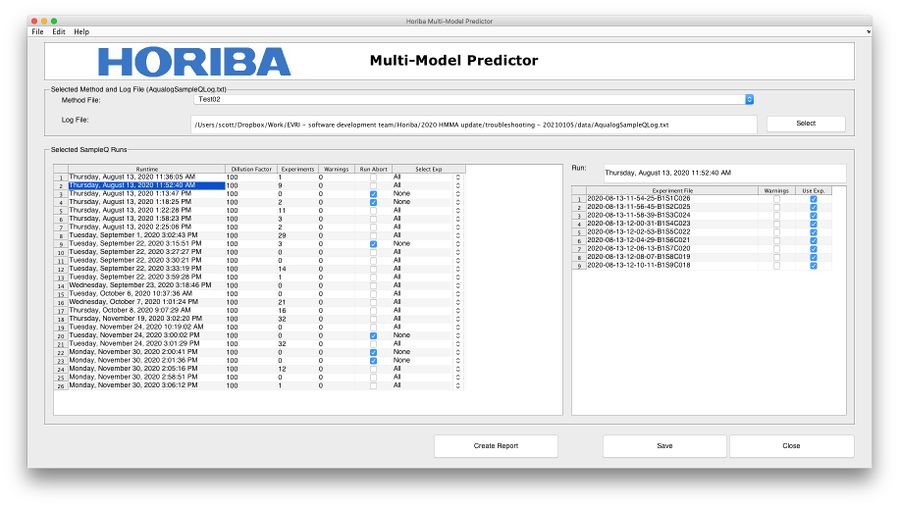
Reporting Interface
Reports can be generated using the following steps:
- If it hasn't been specified already, add a default method file folder using the Edit>Options menu item. Do the same for a default log file location if needed.
- Select a Method from the method drop-down menu.
- Select a log file from a folder containing both the log file and matching data files.
- View Runs in the run table. Experiments will be pre-selected based on abort condition. Use the Select Exp field to automatically adjust experiment selection. Note, right-clicking in the run table will show a context menu that allows for selecting all/no experiments for entire run table.
- Further adjust experiment selection using the experiment table by manually checking the Use Exp. column.
- Click the Create Report button to open a new window with a table of results.
- Click Save from the new window so save a .csv file of the results.
Method Interface
Use these steps to create and edit Method files:
- From the File menu select Admin Login and enter user and password.
- In the new panel the is shown, right-click and select New Method.
- Name the method and, if desired, add a file name.
- Drag and drop a Multi-Block model onto the new method. Note these can be models from within the Solo workspace or as saved .mat files.
- Drag and drop models to apply. For classification models only a single model will be allowed. For regression models, multiple models can be used. Use right-click menu to remove models.
- When a method has been assembled, use the right-click Validate Mehtod menu item to check that the sizes expected by the multiblock model are the same as those expected by the regression/classification models.
- To remove a method, mover or delete the .met file from the parent folder.