Parafac: Difference between revisions
imported>Jeremy (Importing text file) |
m (→Synopsis) |
||
| (53 intermediate revisions by 11 users not shown) | |||
| Line 1: | Line 1: | ||
===Purpose=== | ===Purpose=== | ||
| Line 6: | Line 5: | ||
===Synopsis=== | ===Synopsis=== | ||
:model = parafac(X,'' | :model = parafac(X,ncomp,''options'') | ||
:pred = parafac(Xnew,model) | :pred = parafac(Xnew,model) | ||
:parafac % Launches an analysis window with Parafac as the selected method | |||
Please note that the recommended way to build and apply a PARAFAC model from the command line is to use the Model Object. Please see [[EVRIModel_Objects | this wiki page on building and applying models using the Model Object]]. | |||
===Description=== | ===Description=== | ||
PARAFAC will decompose an array of order ''N'' (where ''N'' | PARAFAC will decompose an array of order ''N'' (where ''N'' >= 3) into the summation over the outer product of ''N'' vectors (a low-rank model). E.g. if ''N''=3 then the array is size ''I'' by ''J'' by ''K''. An example of three-way fluorescence data is shown below.. | ||
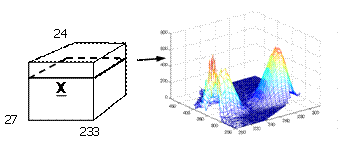
For example, twenty-seven samples containing different amounts of dissolved hydroquinone, tryptophan, phenylalanine, and dopa are measured spectrofluoremetrically using 233 emission wavelengths (250-482 nm) and 24 excitation wavelengths (200-315 nm each 5 nm). A typical sample is also shown. | For example, twenty-seven samples containing different amounts of dissolved hydroquinone, tryptophan, phenylalanine, and dopa are measured spectrofluoremetrically using 233 emission wavelengths (250-482 nm) and 24 excitation wavelengths (200-315 nm each 5 nm). A typical sample is also shown. | ||
[[Image:Parafacdata.gif]] | |||
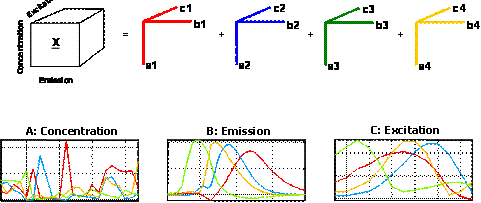
A four-component PARAFAC model of these data will give four factors, each corresponding to one of the chemical analytes. This is illustrated graphically below. The first mode scores (loadings in mode 1) in the matrix '''A''' (27x4) contain estimated relative concentrations of the four analytes in the 27 samples. The second mode loadings '''B''' (233x4) are estimated emission loadings and the third mode loadings '''C''' (24x4) are estimated excitation loadings. | |||
[[Image:Parafacresults.gif]] | |||
For more information about how to use PARAFAC, see the [https://www.youtube.com/playlist?list=PL4L59zaizb3E-Pgp-f90iKHdQQi15JJoL University of Copenhagen's Multi-Way Analysis Videos]. | |||
In the PARAFAC algorithm, any missing values must be set to NaN or Inf and are then automatically handled by expectation maximization. This routine employs an alternating least squares (ALS) algorithm in combination with a line search. For 3-way data, the initial estimate of the loadings is usually obtained from the tri-linear decomposition (TLD). | In the PARAFAC algorithm, any missing values must be set to NaN or Inf and are then automatically handled by expectation maximization. This routine employs an alternating least squares (ALS) algorithm in combination with a line search. For 3-way data, the initial estimate of the loadings is usually obtained from the tri-linear decomposition (TLD). | ||
For assistance in preparing batch data for use in PARAFAC please see [[bspcgui]]. | |||
====Inputs==== | ====Inputs==== | ||
| Line 27: | Line 33: | ||
* '''x''' = the multiway array to be decomposed, and | * '''x''' = the multiway array to be decomposed, and | ||
* '''ncomp''' = the number of factors (components) to use, | * '''ncomp''' = | ||
:* the number of factors (components) to use, OR | |||
* | :* a cell array of parameters such as {a,b,c} which will then be used as starting point for the model. The cell array must be the same length as the number of modes and element j contain the scores/loadings for that mode. If one cell element is empty, this mode is guessed based on the remaining modes. | ||
====Optional Inputs==== | ====Optional Inputs==== | ||
* '''''initval''''' = | * '''''initval''''' = | ||
:* If a parafac model is input, the data are fit to this model where the loadings for the first mode (scores) are estimated. | |||
:* If the loadings are input (e.g. model.loads) these are used as starting values. | |||
*'''''''' | *'''''options''''' = discussed below. | ||
====Outputs==== | ====Outputs==== | ||
| Line 61: | Line 69: | ||
* '''description''': cell array with text description of model, and | * '''description''': cell array with text description of model, and | ||
* '''detail''': sub-structure with additional model details and results. | * '''detail''': sub-structure with additional model details and results. E.g. .detail.ssq contains information on loss function and variance explained as well as variance per component. | ||
Note that the sum-squared captured table contains various statistics on the information captured by each component. Please see [[MCR and PARAFAC Variance Captured]] for details. | |||
The output pred is a structure array that contains the approximation of the data if the options field blockdetails is set to 'all' (see next). | The output pred is a structure array that contains the approximation of the data if the options field blockdetails is set to 'all' (see next). | ||
| Line 75: | Line 84: | ||
* '''weights''': [], used for fitting a weighted loss function (discussed below), | * '''weights''': [], used for fitting a weighted loss function (discussed below), | ||
* ''' | * '''stopcriteria''': Structure defining when to stop iterations based on any one of four criteria | ||
:* '''relativechange''': Default is 1e-6. When the relative change in fit gets below the threshold, the algorithm stops. | |||
:* '''absolutechange''': Default is 1e-6. When the absolute change in fit gets below the threshold, the algorithm stops. | |||
:* '''iterations''': Default is 10.000. When the number of iterations exceeds the threshold, the algorithm stops. | |||
:* '''seconds''': Default is 3600 (seconds). When the time spent exceeds the threshold, the algorithm stops. | |||
* '''init''': [ 0 ], defines how parameters are initialized (discussed below), | * '''init''': [ 0 ], defines how parameters are initialized (discussed below), | ||
| Line 81: | Line 95: | ||
* '''line''': [ 0 | {1}] defines whether to use the line search {default uses it}, | * '''line''': [ 0 | {1}] defines whether to use the line search {default uses it}, | ||
* '''algo''': [ {'ALS'} | 'tld' | 'swatld' ] governs algorithm used, | * '''algo''': [ {'ALS'} | 'tld' | 'swatld' ] governs algorithm used. Only ALS allows more than three-way and allows constraints, | ||
* '''iterative''': settings for iterative reweighted least squares fitting (see help on weights below), | * '''iterative''': settings for iterative reweighted least squares fitting (see help on weights below), | ||
* ''' | * '''validation.splithalf''': [ 'on' | {'off'} ], Allows doing [[splithalf]] analysis. See the help of SPLITHALF for more information, | ||
* '''auto_outlier.perform''': [ 'on' | {'off'} ], Will automatically remove detected outliers in an iterative fashion. See auto_outlier.help for more information, | |||
* ''' | * '''scaletype''': Defines how loadings are scaled. See options.scaletype.text for help, | ||
* '''blockdetails''': [ {'standard'} | 'compact' | 'all' ] level of detail (predictions, raw residuals, and calibration data) included in the model. | |||
:* ‘Standard’ = the predictions and raw residuals for the X-block as well as the X-block itself are not stored in the model to reduce its size in memory. Specifically, these fields in the model object are left empty: 'model.pred{1}', 'model.detail.res{1}', 'model.detail.data{1}'. | |||
:* ‘Compact’ = like 'Standard' only residual limits from old model is used and the core consistency field in the model structure is left empty. ('model.detail.reslim', 'model.detail.coreconsistency.consistency'). | |||
:* 'All' = keep predictions, raw residuals for x-block as well as the X-block dataset itself. | |||
* '''preprocessing''': {[]}, one element cell array containing preprocessing structure (see PREPROCESS) defining preprocessing to use on the x-block | |||
* '''samplemode''': [1], defines which mode should be considered the sample or object mode, | * '''samplemode''': [1], defines which mode should be considered the sample or object mode, | ||
* '''constraints''': {3x1 cell}, defines constraints on parameters (discussed below), and | * '''constraints''': {3x1 cell}, defines constraints on parameters (discussed below), | ||
* '''coreconsist''': [ {'on'} | 'off' ], governs calculation of core consistency (turning off may save time with large data sets and many components), and | |||
* ''' | * '''waitbar''': [ {'on'} | 'off' ], display waitbar. | ||
The default options can be retrieved using: options = parafac('options');. | The default options can be retrieved using: options = parafac('options');. | ||
=====Weights===== | |||
Through the use of the ''options'' field weights it is possible to fit a PARAFAC model in a weighted least squares sense The input is an array of the same size as the input data X holding individual weights for each element. The PARAFAC model is then fit in a weighted least squares sense. Instead of minimizing the frobenius norm ||x-M||<sup>2</sup> where M is the PARAFAC model, the norm ||(x-M). | Through the use of the ''options'' field weights it is possible to fit a PARAFAC model in a weighted least squares sense The input is an array of the same size as the input data X holding individual weights for each element. The PARAFAC model is then fit in a weighted least squares sense. Instead of minimizing the frobenius norm ||x-M||<sup>2</sup> where M is the PARAFAC model, the norm ||(x-M).*weights||<sup>2</sup> is minimized. The algorithm used for weighted regression is based on a majorization step according to Kiers, ''Psychometrika'', '''62''', 251-266, 1997 which has the advantage of being computationally inexpensive. | ||
=====Init===== | |||
The ''options'' field init is used to govern how the initial guess for the loadings is obtained. If optional input ''initval'' is input then options.init is not used. The following choices for init are available. | The ''options'' field init is used to govern how the initial guess for the loadings is obtained. If optional input ''initval'' is input then options.init is not used. The following choices for init are available. | ||
| Line 109: | Line 134: | ||
* '''init''' = 0, PARAFAC chooses initialization {default}, | * '''init''' = 0, PARAFAC chooses initialization {default}, | ||
* '''init''' = 1, uses TLD (unless | * '''init''' = 1, uses TLD (unless data is more than three-way. Then ATLD is used), | ||
* '''init''' = 2, | * '''init''' = 2, based on singular value decomposition (good alternative to 1), | ||
* '''init''' = 3, based on orthogonalization of random values ( | * '''init''' = 3, based on orthogonalization of random values (good for checking local minima), | ||
* '''init''' = 4, based on | * '''init''' = 4, based on approximate (sequentially fitted) PARAFAC model, | ||
* '''init''' = 5, based on compression which may be useful for large data, and | * '''init''' = 5, based on compression which may be useful for large data, and | ||
| Line 121: | Line 146: | ||
* '''init''' > 5, based on best fit of many (the value options.init) small runs. | * '''init''' > 5, based on best fit of many (the value options.init) small runs. | ||
=====Constraints===== | |||
The ''options'' field constraints is used to employ constraints on the parameters. It is a cell array with number of elements equal to the number of modes of the input data X. Each cell contains a structure array that defines the constraints in that particular mode. Hence, options.constraints{2} defines constraints on the second mode loadings. For help on setting constraints see [[constrainfit]]. Note, that if your dataset is e.g. a five-way array, then the default constraint field in options only defines the first three modes. You will have to make the constraint field for the remaining modes yourself. This can be done by copying from the other modes. For example, options.constraints{4} = options.constraints{1};options.constraints{5} = options.constraints{1}; | |||
===Examples=== | ===Examples=== | ||
| Line 183: | Line 170: | ||
===See Also=== | ===See Also=== | ||
[[datahat]], [[explode]], [[gram]], [[mpca]], [[outerm]], [[parafac2]], [[tld]], [[tucker]], [[unfoldm]] | [[analysis]], [[bspcgui]], [[datahat]], [[eemoutlier]], [[explode]], [[gram]], [[mpca]], [[npls]], [[outerm]], [[parafac2]], [[pca]], [[preprocess]], [[splithalf]], [[tld]], [[tucker]], [[unfoldm]], [[modelviewer]], [[EVRIModel_Objects]] | ||
Latest revision as of 23:31, 12 November 2020
Purpose
PARAFAC (PARAllel FACtor analysis) for multi-way arrays
Synopsis
- model = parafac(X,ncomp,options)
- pred = parafac(Xnew,model)
- parafac % Launches an analysis window with Parafac as the selected method
Please note that the recommended way to build and apply a PARAFAC model from the command line is to use the Model Object. Please see this wiki page on building and applying models using the Model Object.
Description
PARAFAC will decompose an array of order N (where N >= 3) into the summation over the outer product of N vectors (a low-rank model). E.g. if N=3 then the array is size I by J by K. An example of three-way fluorescence data is shown below..
For example, twenty-seven samples containing different amounts of dissolved hydroquinone, tryptophan, phenylalanine, and dopa are measured spectrofluoremetrically using 233 emission wavelengths (250-482 nm) and 24 excitation wavelengths (200-315 nm each 5 nm). A typical sample is also shown.
A four-component PARAFAC model of these data will give four factors, each corresponding to one of the chemical analytes. This is illustrated graphically below. The first mode scores (loadings in mode 1) in the matrix A (27x4) contain estimated relative concentrations of the four analytes in the 27 samples. The second mode loadings B (233x4) are estimated emission loadings and the third mode loadings C (24x4) are estimated excitation loadings.
For more information about how to use PARAFAC, see the University of Copenhagen's Multi-Way Analysis Videos.
In the PARAFAC algorithm, any missing values must be set to NaN or Inf and are then automatically handled by expectation maximization. This routine employs an alternating least squares (ALS) algorithm in combination with a line search. For 3-way data, the initial estimate of the loadings is usually obtained from the tri-linear decomposition (TLD).
For assistance in preparing batch data for use in PARAFAC please see bspcgui.
Inputs
- x = the multiway array to be decomposed, and
- ncomp =
- the number of factors (components) to use, OR
- a cell array of parameters such as {a,b,c} which will then be used as starting point for the model. The cell array must be the same length as the number of modes and element j contain the scores/loadings for that mode. If one cell element is empty, this mode is guessed based on the remaining modes.
Optional Inputs
- initval =
- If a parafac model is input, the data are fit to this model where the loadings for the first mode (scores) are estimated.
- If the loadings are input (e.g. model.loads) these are used as starting values.
- options = discussed below.
Outputs
The output model is a structure array with the following fields:
- modeltype: 'PARAFAC',
- datasource: structure array with information about input data,
- date: date of creation,
- time: time of creation,
- info: additional model information,
- loads: 1 by K cell array with model loadings for each mode/dimension,
- pred: cell array with model predictions for each input data block,
- tsqs: cell array with T2 values for each mode,
- ssqresiduals: cell array with sum of squares residuals for each mode,
- description: cell array with text description of model, and
- detail: sub-structure with additional model details and results. E.g. .detail.ssq contains information on loss function and variance explained as well as variance per component.
Note that the sum-squared captured table contains various statistics on the information captured by each component. Please see MCR and PARAFAC Variance Captured for details. The output pred is a structure array that contains the approximation of the data if the options field blockdetails is set to 'all' (see next).
Options
options = a structure array with the following fields:
- display: [ {'on'} | 'off' ], governs level of display,
- plots: [ {'final'} | 'all' | 'none' ], governs level of plotting,
- weights: [], used for fitting a weighted loss function (discussed below),
- stopcriteria: Structure defining when to stop iterations based on any one of four criteria
- relativechange: Default is 1e-6. When the relative change in fit gets below the threshold, the algorithm stops.
- absolutechange: Default is 1e-6. When the absolute change in fit gets below the threshold, the algorithm stops.
- iterations: Default is 10.000. When the number of iterations exceeds the threshold, the algorithm stops.
- seconds: Default is 3600 (seconds). When the time spent exceeds the threshold, the algorithm stops.
- init: [ 0 ], defines how parameters are initialized (discussed below),
- line: [ 0 | {1}] defines whether to use the line search {default uses it},
- algo: [ {'ALS'} | 'tld' | 'swatld' ] governs algorithm used. Only ALS allows more than three-way and allows constraints,
- iterative: settings for iterative reweighted least squares fitting (see help on weights below),
- validation.splithalf: [ 'on' | {'off'} ], Allows doing splithalf analysis. See the help of SPLITHALF for more information,
- auto_outlier.perform: [ 'on' | {'off'} ], Will automatically remove detected outliers in an iterative fashion. See auto_outlier.help for more information,
- scaletype: Defines how loadings are scaled. See options.scaletype.text for help,
- blockdetails: [ {'standard'} | 'compact' | 'all' ] level of detail (predictions, raw residuals, and calibration data) included in the model.
- ‘Standard’ = the predictions and raw residuals for the X-block as well as the X-block itself are not stored in the model to reduce its size in memory. Specifically, these fields in the model object are left empty: 'model.pred{1}', 'model.detail.res{1}', 'model.detail.data{1}'.
- ‘Compact’ = like 'Standard' only residual limits from old model is used and the core consistency field in the model structure is left empty. ('model.detail.reslim', 'model.detail.coreconsistency.consistency').
- 'All' = keep predictions, raw residuals for x-block as well as the X-block dataset itself.
- preprocessing: {[]}, one element cell array containing preprocessing structure (see PREPROCESS) defining preprocessing to use on the x-block
- samplemode: [1], defines which mode should be considered the sample or object mode,
- constraints: {3x1 cell}, defines constraints on parameters (discussed below),
- coreconsist: [ {'on'} | 'off' ], governs calculation of core consistency (turning off may save time with large data sets and many components), and
- waitbar: [ {'on'} | 'off' ], display waitbar.
The default options can be retrieved using: options = parafac('options');.
Weights
Through the use of the options field weights it is possible to fit a PARAFAC model in a weighted least squares sense The input is an array of the same size as the input data X holding individual weights for each element. The PARAFAC model is then fit in a weighted least squares sense. Instead of minimizing the frobenius norm ||x-M||2 where M is the PARAFAC model, the norm ||(x-M).*weights||2 is minimized. The algorithm used for weighted regression is based on a majorization step according to Kiers, Psychometrika, 62, 251-266, 1997 which has the advantage of being computationally inexpensive.
Init
The options field init is used to govern how the initial guess for the loadings is obtained. If optional input initval is input then options.init is not used. The following choices for init are available.
Generally, options.init = 0, will do for well-behaved data whereas options.init = 10, will be suitable for difficult models. Difficult models are typically those with many components, with very correlated loadings, or models where there are indications that local minima are present.
- init = 0, PARAFAC chooses initialization {default},
- init = 1, uses TLD (unless data is more than three-way. Then ATLD is used),
- init = 2, based on singular value decomposition (good alternative to 1),
- init = 3, based on orthogonalization of random values (good for checking local minima),
- init = 4, based on approximate (sequentially fitted) PARAFAC model,
- init = 5, based on compression which may be useful for large data, and
- init > 5, based on best fit of many (the value options.init) small runs.
Constraints
The options field constraints is used to employ constraints on the parameters. It is a cell array with number of elements equal to the number of modes of the input data X. Each cell contains a structure array that defines the constraints in that particular mode. Hence, options.constraints{2} defines constraints on the second mode loadings. For help on setting constraints see constrainfit. Note, that if your dataset is e.g. a five-way array, then the default constraint field in options only defines the first three modes. You will have to make the constraint field for the remaining modes yourself. This can be done by copying from the other modes. For example, options.constraints{4} = options.constraints{1};options.constraints{5} = options.constraints{1};
Examples
parafac demo gives a demonstration of the use of the PARAFAC algorithm.
model = parafac(X,5) fits a five-component PARAFAC model to the array X using default settings.
pred = parafac(Z,model) fits a parafac model to new data Z. The scores will be taken to be in the first mode, but you can change this by setting options.samplemodex to the mode which is the sample mode. Note, that the sample-mode dimension may be different for the old model and the new data, but all other dimensions must be the same.
options = parafac('options'); generates a set of default settings for PARAFAC. options.plots = 0; sets the plotting off.
options.init = 3; sets the initialization of PARAFAC to orthogonalized random numbers.
options.samplemodex = 2; Defines the second mode to be the sample-mode. Useful, for example, when fitting an existing model to new data has to provide the scores in the second mode.
model = parafac(X,2,options); fits a two-component PARAFAC model with the settings defined in options.
parafac io shows the I/O of the algorithm.
See Also
analysis, bspcgui, datahat, eemoutlier, explode, gram, mpca, npls, outerm, parafac2, pca, preprocess, splithalf, tld, tucker, unfoldm, modelviewer, EVRIModel_Objects